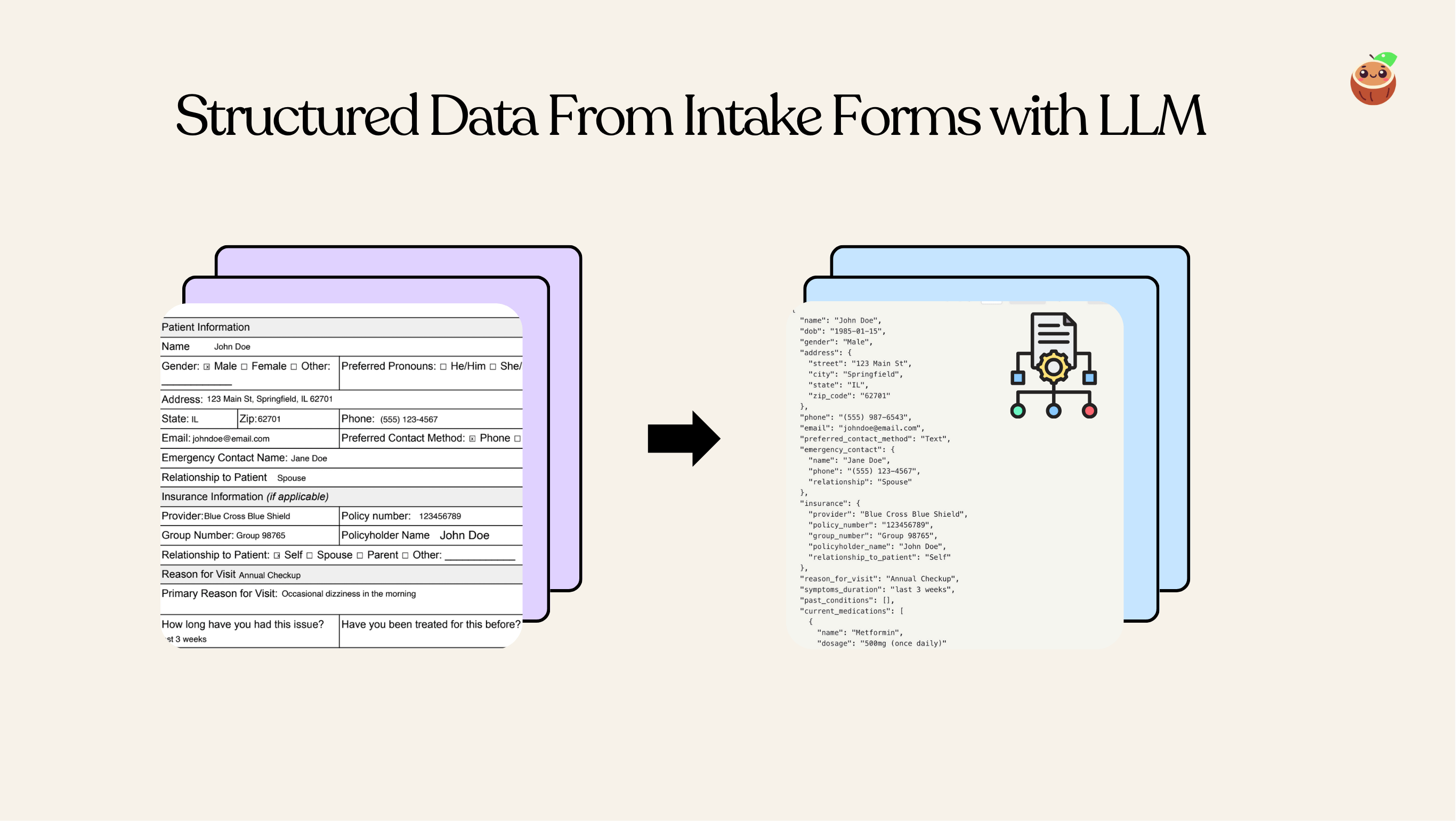
Extract Nested Structured Data from Patient Form
Overview
With CocoIndex, you can easily define nested schema in Python dataclass and use LLM to extract structured data from unstructured data. This example shows how to extract structured data from patient intake forms.
The extraction quality is highly dependent on the OCR quality. You can use CocoIndex with any commercial parser or open source ones that is tailored for your domain for better results. For example, Document AI from Google Cloud and more.
Flow Overview
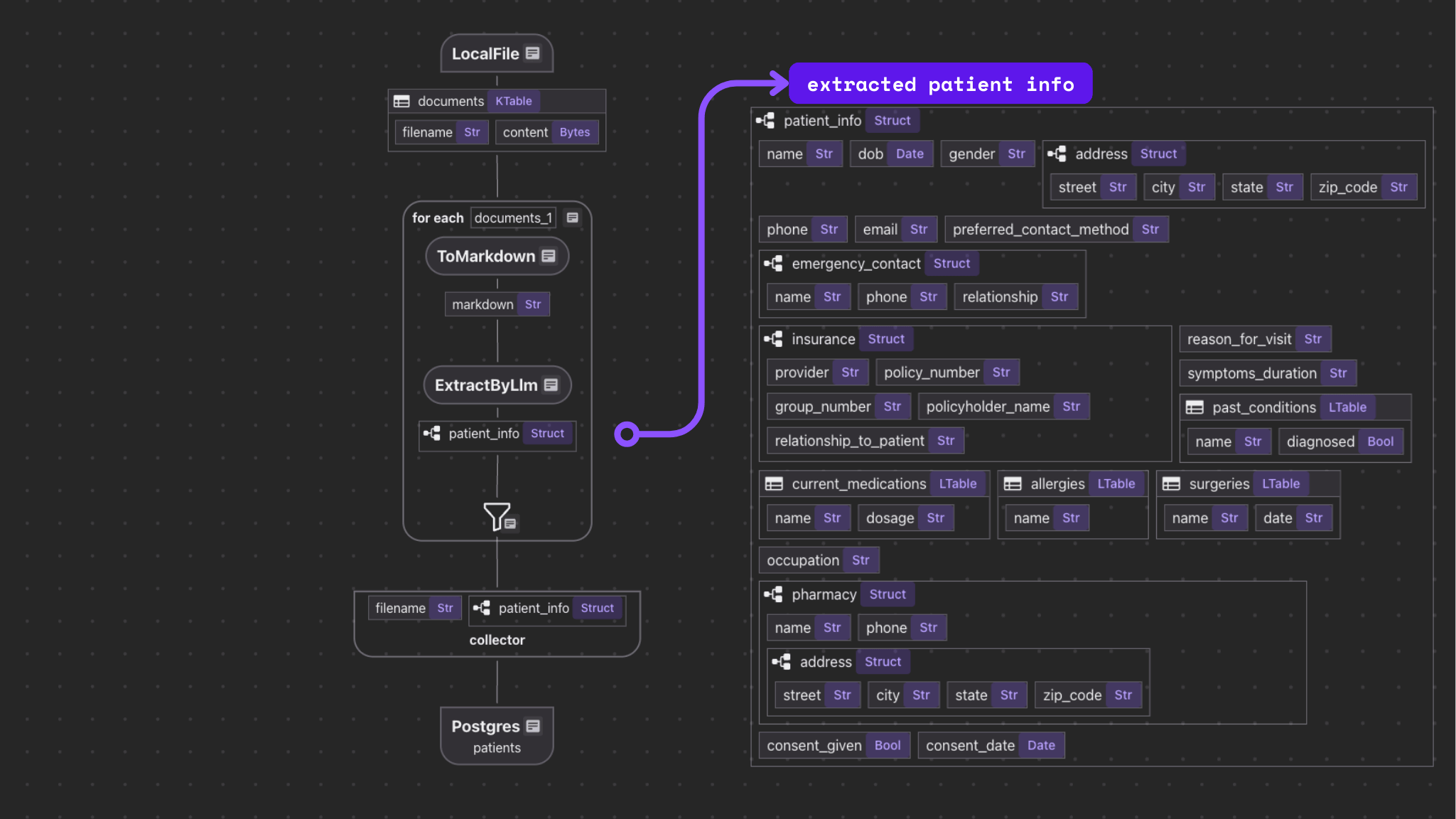
The flow itself is fairly simple.
- Import a list o intake forms.
- For each file:
- Convert the file to Markdown.
- Extract structured data from the Markdown.
- Export selected fields to tables in Postgres with PGVector.
Setup
- If you don't have Postgres installed, please refer to the installation guide.
- Configure your OpenAI API key. Create a
.envfile from.env.example, and fillOPENAI_API_KEY.
Alternatively, we have native support for Gemini, Ollama, LiteLLM. You can choose your favorite LLM provider and work completely on-premises.
LLMAdd source
Add source from local files.
import os
@cocoindex.flow_def(name="PatientIntakeExtraction")
def patient_intake_extraction_flow(
flow_builder: cocoindex.FlowBuilder, data_scope: cocoindex.DataScope
):
"""
Define a flow that extracts patient information from intake forms.
"""
data_scope["documents"] = flow_builder.add_source(
cocoindex.sources.LocalFile(path=os.path.join('data', 'patient_forms'), binary=True)
)
flow_builder.add_source will create a table with a few sub fields.
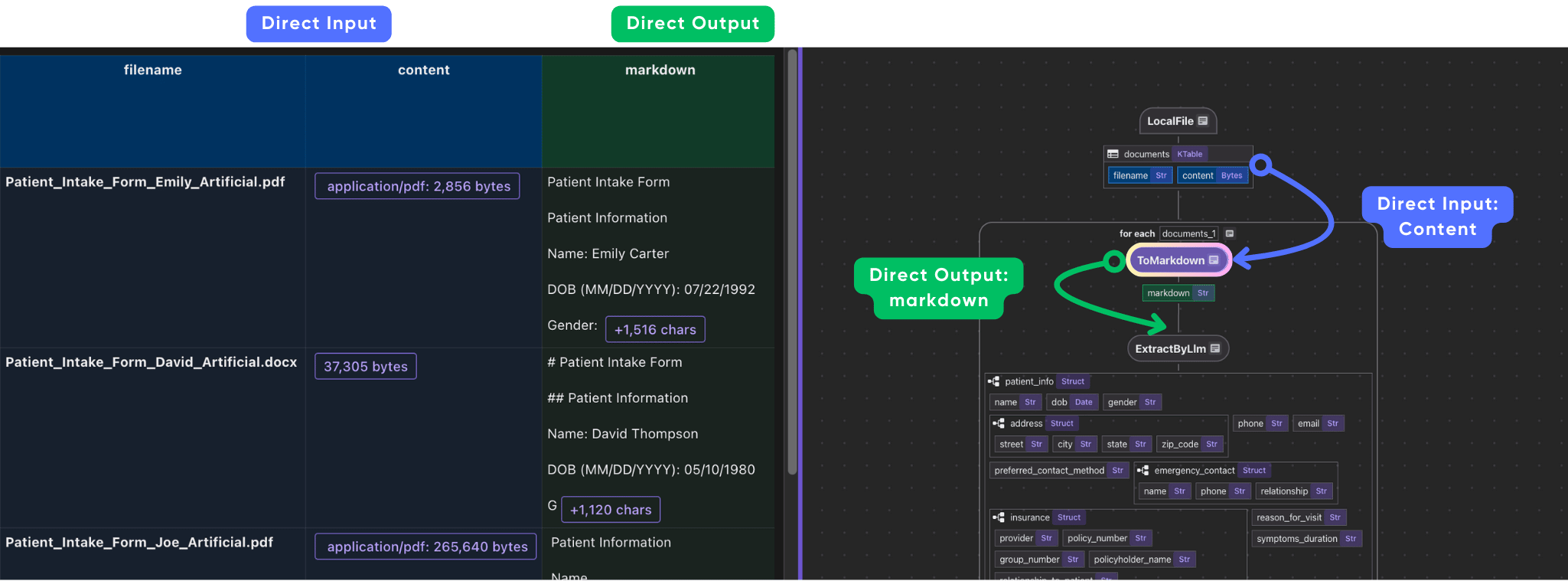
Parse documents with different formats to Markdown
Define a custom function to parse documents in any format to Markdown. Here we use MarkItDown to convert the file to Markdown. It also provides options to parse by LLM, like gpt-4o. At present, MarkItDown supports: PDF, Word, Excel, Images (EXIF metadata and OCR), etc.
class ToMarkdown(cocoindex.op.FunctionSpec):
"""Convert a document to markdown."""
@cocoindex.op.executor_class(gpu=True, cache=True, behavior_version=1)
class ToMarkdownExecutor:
"""Executor for ToMarkdown."""
spec: ToMarkdown
_converter: MarkItDown
def prepare(self):
client = OpenAI()
self._converter = MarkItDown(llm_client=client, llm_model="gpt-4o")
def __call__(self, content: bytes, filename: str) -> str:
suffix = os.path.splitext(filename)[1]
with tempfile.NamedTemporaryFile(delete=True, suffix=suffix) as temp_file:
temp_file.write(content)
temp_file.flush()
text = self._converter.convert(temp_file.name).text_content
return text
Next we plug it into the data flow.
with data_scope["documents"].row() as doc:
doc["markdown"] = doc["content"].transform(ToMarkdown(), filename=doc["filename"])
Define output schema
We are going to define the patient info schema for structured extraction. One of the best examples to define a patient info schema is probably following the FHIR standard - Patient Resource.
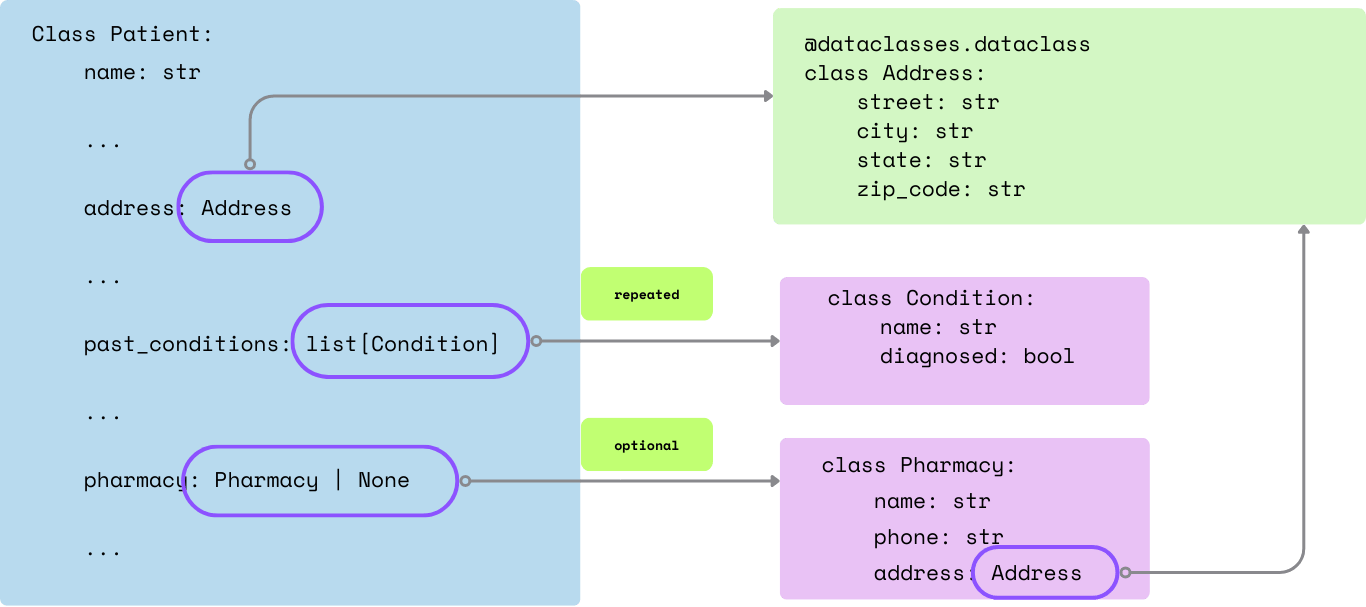
In this tutorial, we'll define a simplified schema in nested dataclass for patient information extraction:
@dataclasses.dataclass
class Contact:
name: str
phone: str
relationship: str
@dataclasses.dataclass
class Address:
street: str
city: str
state: str
zip_code: str
@dataclasses.dataclass
class Pharmacy:
name: str
phone: str
address: Address
@dataclasses.dataclass
class Insurance:
provider: str
policy_number: str
group_number: str | None
policyholder_name: str
relationship_to_patient: str
@dataclasses.dataclass
class Condition:
name: str
diagnosed: bool
@dataclasses.dataclass
class Medication:
name: str
dosage: str
@dataclasses.dataclass
class Allergy:
name: str
@dataclasses.dataclass
class Surgery:
name: str
date: str
@dataclasses.dataclass
class Patient:
name: str
dob: datetime.date
gender: str
address: Address
phone: str
email: str
preferred_contact_method: str
emergency_contact: Contact
insurance: Insurance | None
reason_for_visit: str
symptoms_duration: str
past_conditions: list[Condition]
current_medications: list[Medication]
allergies: list[Allergy]
surgeries: list[Surgery]
occupation: str | None
pharmacy: Pharmacy | None
consent_given: bool
consent_date: datetime.date | None
A simplified illustration of the nested fields and its definition:
Extract structured data from Markdown
CocoIndex provides built-in functions (e.g. ExtractByLlm) that process data using LLMs. With CocoIndex, you can directly pass the Python dataclass Patient to the function, and it will automatically parse the LLM response into the dataclass.
with data_scope["documents"].row() as doc:
doc["patient_info"] = doc["markdown"].transform(
cocoindex.functions.ExtractByLlm(
llm_spec=cocoindex.LlmSpec(
api_type=cocoindex.LlmApiType.OPENAI, model="gpt-4o"),
output_type=Patient,
instruction="Please extract patient information from the intake form."))
patients_index.collect(
filename=doc["filename"],
patient_info=doc["patient_info"],
)
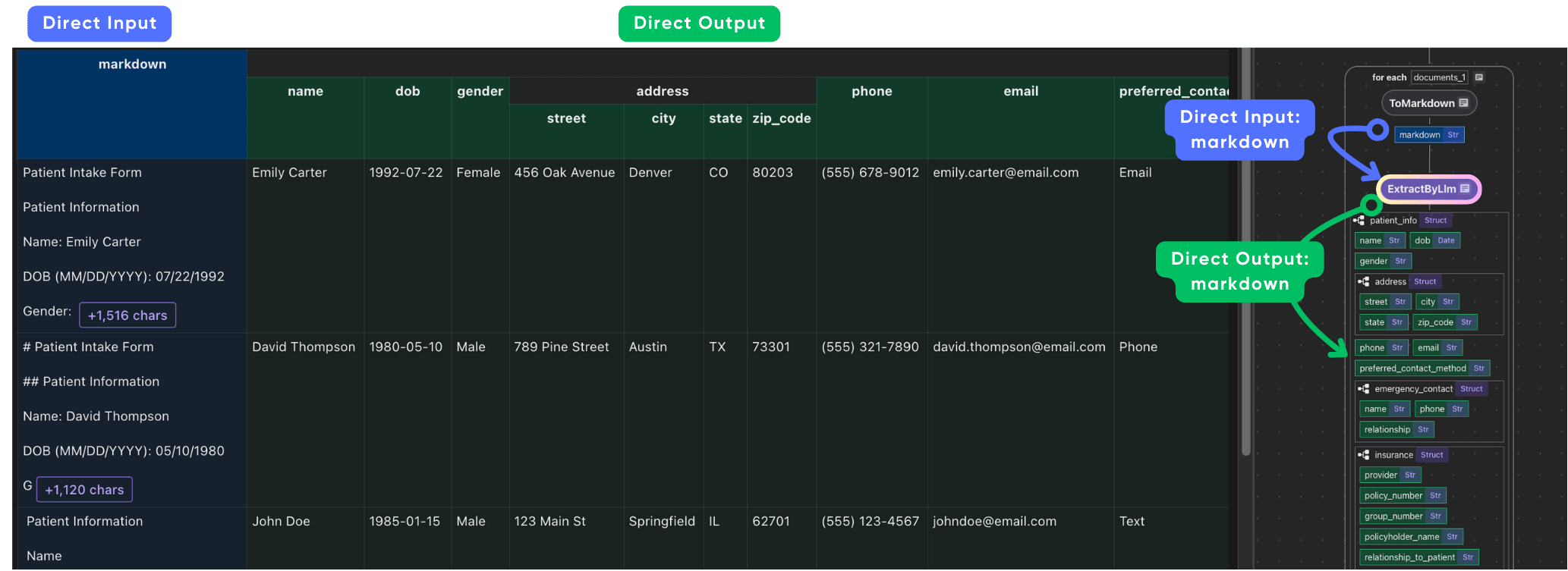
After the extraction, we collect all the fields for simplicity. You can also select any fields and also perform data mapping and field level transformation on the fields before the collection.
Export the extracted data to a table
patients_index.export(
"patients",
cocoindex.storages.Postgres(table_name="patients_info"),
primary_key_fields=["filename"],
)
Run and Query
Install dependencies
pip install -e .
Setup and update the index
cocoindex update main
You'll see the index updates state in the terminal
Query the output table
After the index is built, you have a table with the name patients_info. You can query it at any time, e.g., start a Postgres shell:
psql postgres://cocoindex:cocoindex@localhost/cocoindex
The run:
select * from patients_info;
You could see the patients_info table.
Evaluate
For mission-critical use cases, it is important to evaluate the quality of the extraction. CocoIndex supports a simple way to evaluate the extraction. More updates are coming soon.
-
Dump the extracted data to YAML files.
python3 main.py cocoindex evaluateIt dumps what should be indexed to files under a directory. Using my example data sources, it looks like the golden files with a timestamp on the directory name.
-
Compare the extracted data with golden files. We created a directory with golden files for each patient intake form. You can find them here.
You can run the following command to see the diff:
diff -r data/eval_PatientIntakeExtraction_golden data/eval_PatientIntakeExtraction_outputI used a tool called DirEqual for mac. We also recommend Meld for Linux and Windows.
A diff from DirEqual looks like this:
And double click on any row to see file level diff. In my case, there's missing
conditionforPatient_Intake_Form_Joe.pdffile.
Troubleshooting
If extraction is not ideal, this is how I troubleshoot. My original golden file for this record is this one.
We could troubleshoot in two steps:
- Convert to Markdown
- Extract structured data from Markdown
I also use CocoInsight to help me troubleshoot.
cocoindex server -ci main
Go to https://cocoindex.io/cocoinsight. You could see an interactive UI to explore the data.
Click on the markdown column for Patient_Intake_Form_Joe.pdf, you could see the Markdown content. We could try a few different models with the Markdown converter/LLM to iterate and see if we can get better results, or needs manual correction.
Connect to other sources
CocoIndex natively supports Google Drive, Amazon S3, Azure Blob Storage, and more.
Sources