Index academic papers and extract metadata for AI agents
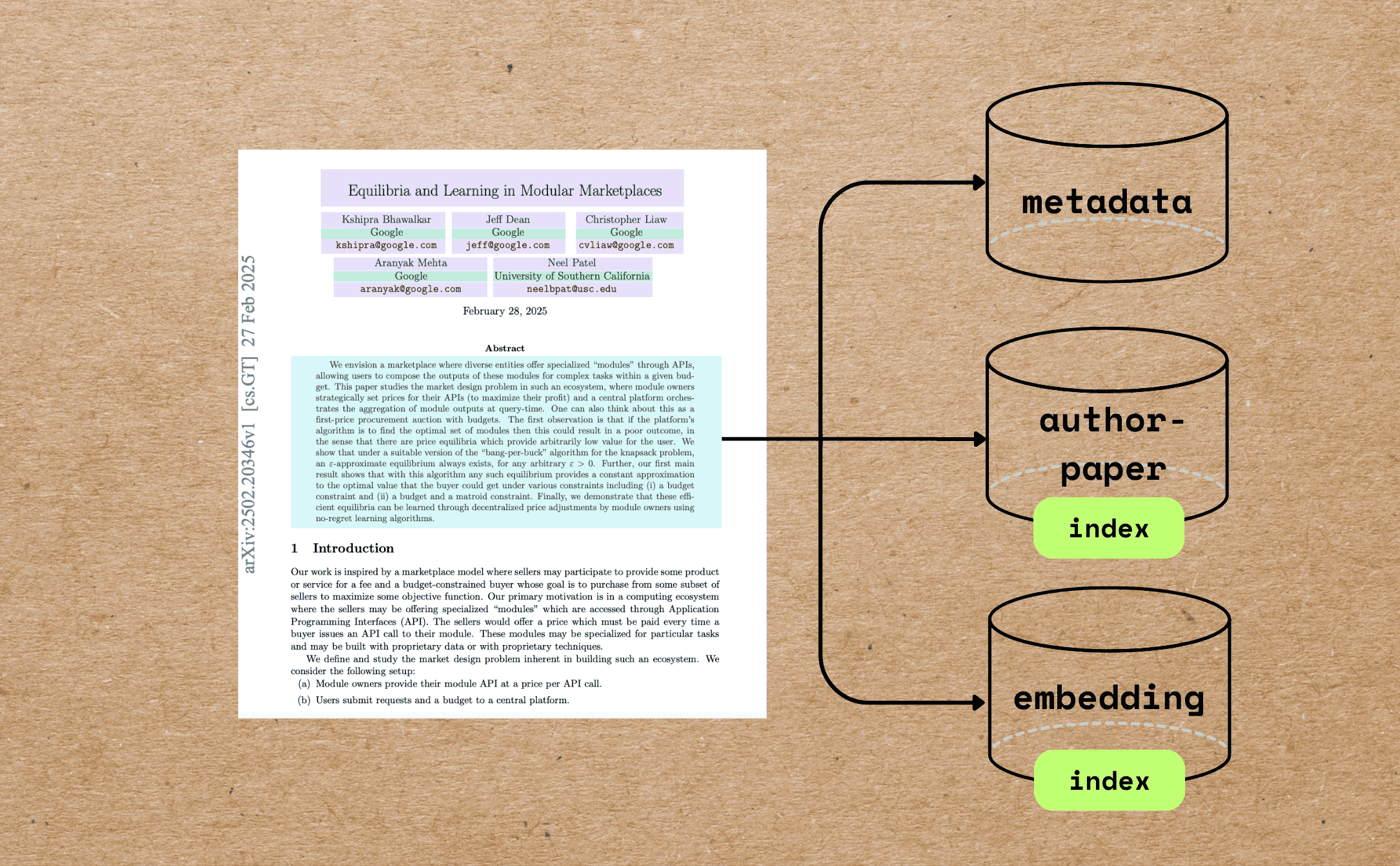
In this blog we will walk through a comprehensive example of indexing research papers with extracting different metadata — beyond full text chunking and embedding — and build semantic embeddings for indexing and querying.
We would greatly appreciate it if you could ⭐ star CocoIndex on GitHub if you find this tutorial helpful.
Use cases
- Academic search and retrieval, as well as research-based AI agents
- Paper recommendation systems
- Research knowledge graphs
- Semantic analysis of scientific literature
What we will achieve
Let's take a look at this PDF as an example.
Here’s what we want to accomplish:
-
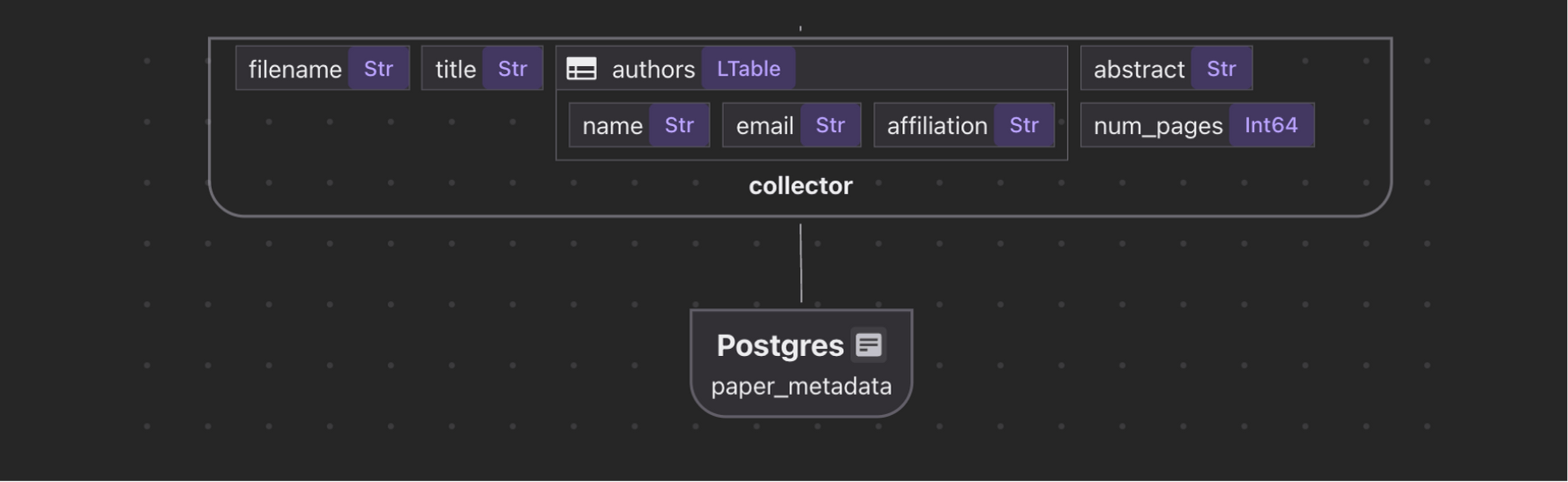
Extract the paper metadata, including file name, title, author information, abstract, and number of pages.
-
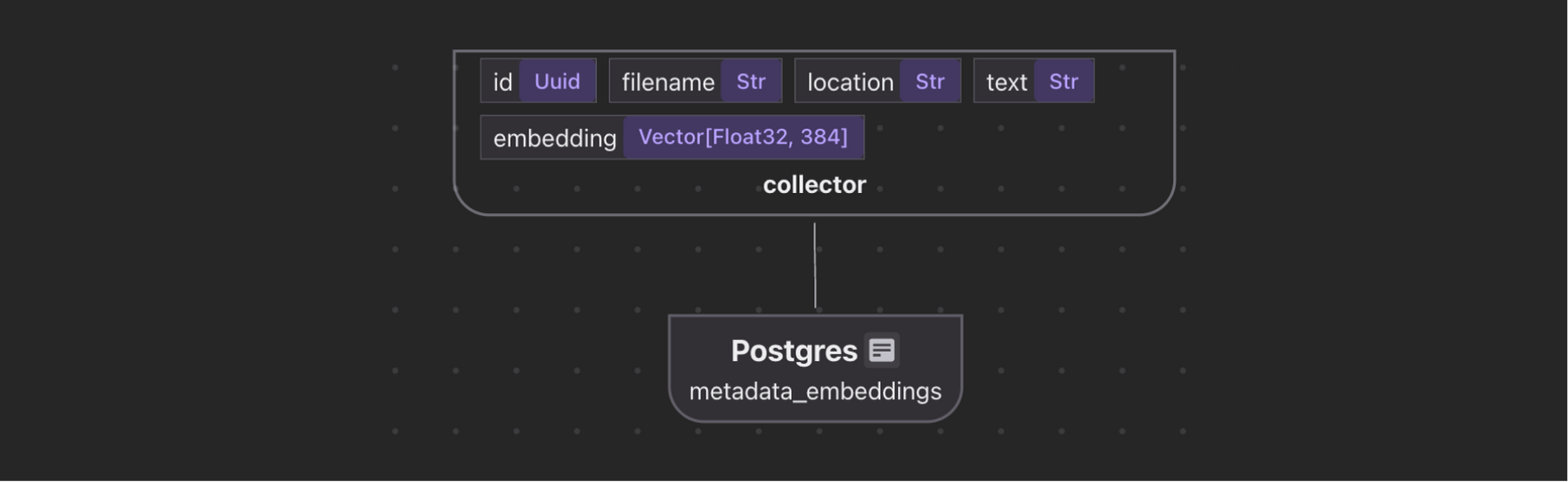
Build vector embeddings for the metadata, such as the title and abstract, for semantic search. This enables better metadata-driven semantic search results. For example, you can match text queries against titles and abstracts.
-
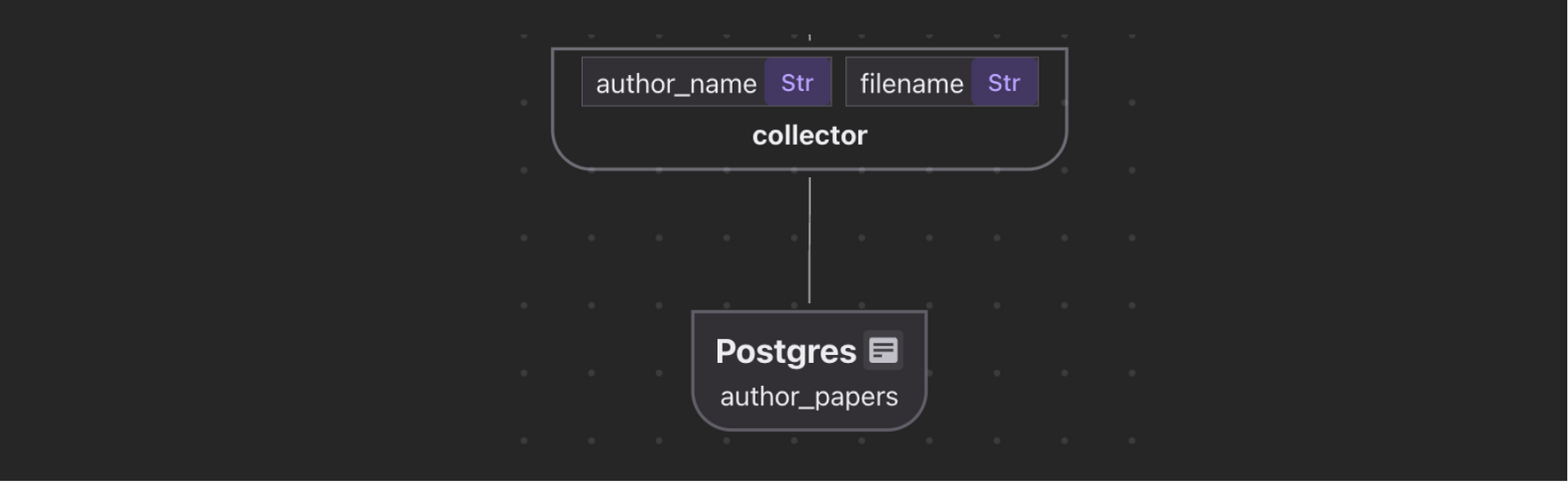
Build an index of authors and all the file names associated with each author to answer questions like "Give me all the papers by Jeff Dean."
-
If you want to perform full PDF embedding for the paper, you can also refer to this article.
You can find the full code here.
Core components
-
PDF Preprocessing
- Reads PDFs using
pypdfand extracts:- Total number of pages
- First page content (used as a proxy for metadata-rich information)
- Reads PDFs using
-
Markdown Conversion
- Converts the first page to Markdown using Marker.
-
LLM-Powered Metadata Extraction
- Sends the first-page Markdown to GPT-4o using CocoIndex's
ExtractByLlmfunction. - Extracted metadata includes and more.
title(string)authors(with name, email, and affiliation)abstract(string)
- Sends the first-page Markdown to GPT-4o using CocoIndex's
-
Semantic Embedding
- The title is embedded directly using the
all-MiniLM-L6-v2model by the SentenceTransformer. - Abstracts are chunked based on semantic punctuation and token count, then each chunk is embedded individually.
- The title is embedded directly using the
-
Relational Data Collection
- Authors are unrolled and collected into an
author_papersrelation, enabling queries like:- Show all papers by X
- Which co-authors worked with Y?
- Authors are unrolled and collected into an
Prerequisites
- Install PostgreSQL. CocoIndex uses PostgreSQL internally for incremental processing.
- Configure your OpenAI API key. Alternatively, we have native support for Gemini, Ollama, LiteLLM, checkout the guide. You could choose your favorite LLM provider and can work completely on-premises.
Define indexing flow
This project demonstrates a slightly more comprehensive example of metadata understanding closer to real-world use cases. You’ll see how easy it is to achieve this design by CocoIndex within 100 lines of indexing logic - code.
To better help you navigate what we will walk through, here is a flow diagram.
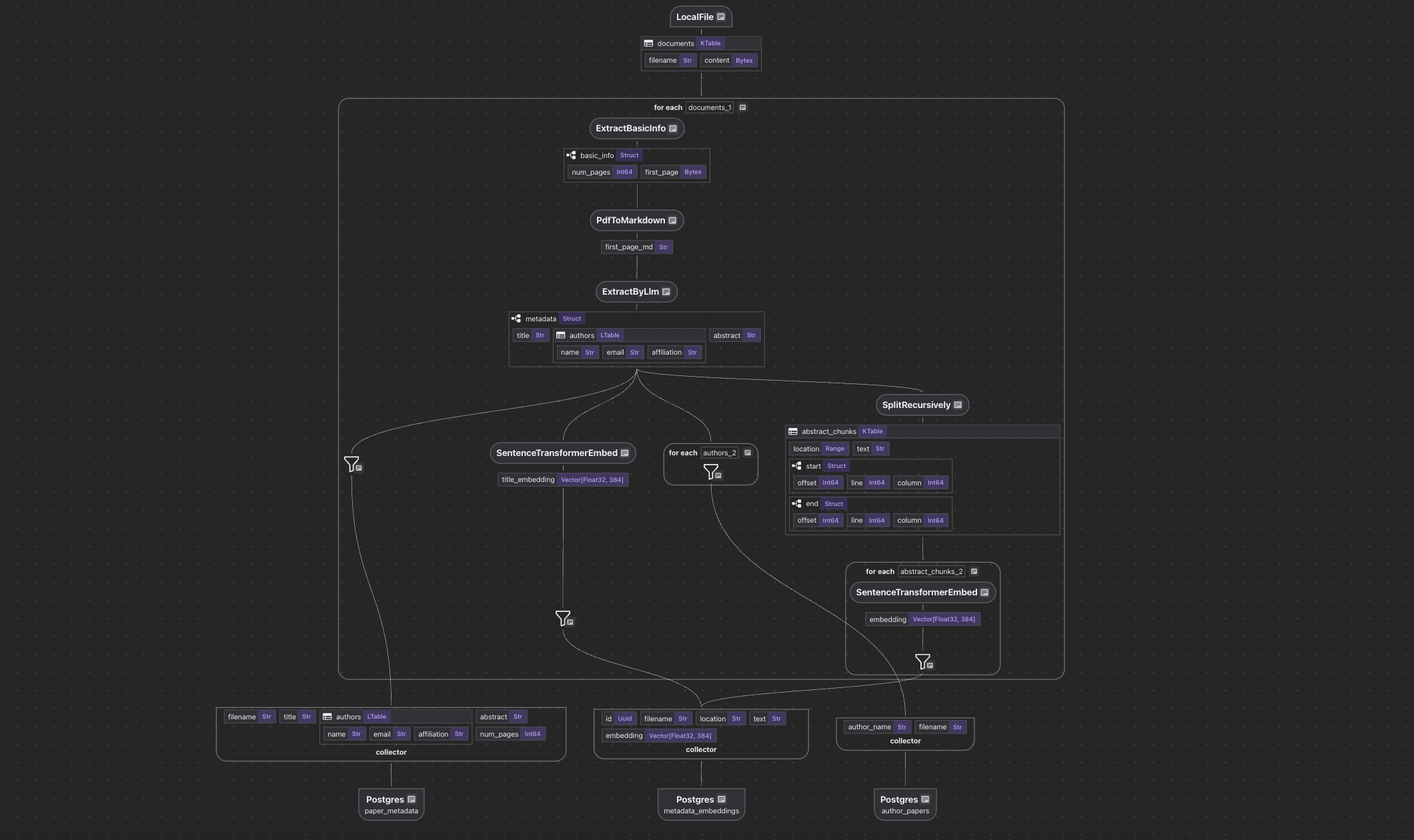
- Import a list of papers in PDF.
- For each file:
- Extract the first page of the paper.
- Convert the first page to Markdown.
- Extract metadata (title, authors, abstract) from the first page.
- Split the abstract into chunks, and compute embeddings for each chunk.
- Export to the following tables in Postgres with PGVector:
- Metadata (title, authors, abstract) for each paper.
- Author-to-paper mapping, for author-based query.
- Embeddings for titles and abstract chunks, for semantic search.
Let’s zoom in the steps.
Import the papers
@cocoindex.flow_def(name="PaperMetadata")
def paper_metadata_flow(
flow_builder: cocoindex.FlowBuilder, data_scope: cocoindex.DataScope
) -> None:
data_scope["documents"] = flow_builder.add_source(
cocoindex.sources.LocalFile(path="papers", binary=True),
refresh_interval=datetime.timedelta(seconds=10),
)
flow_builder.add_source will create a table with sub fields (filename, content),
we can refer to the documentation for more details.

Extract and collect metadata
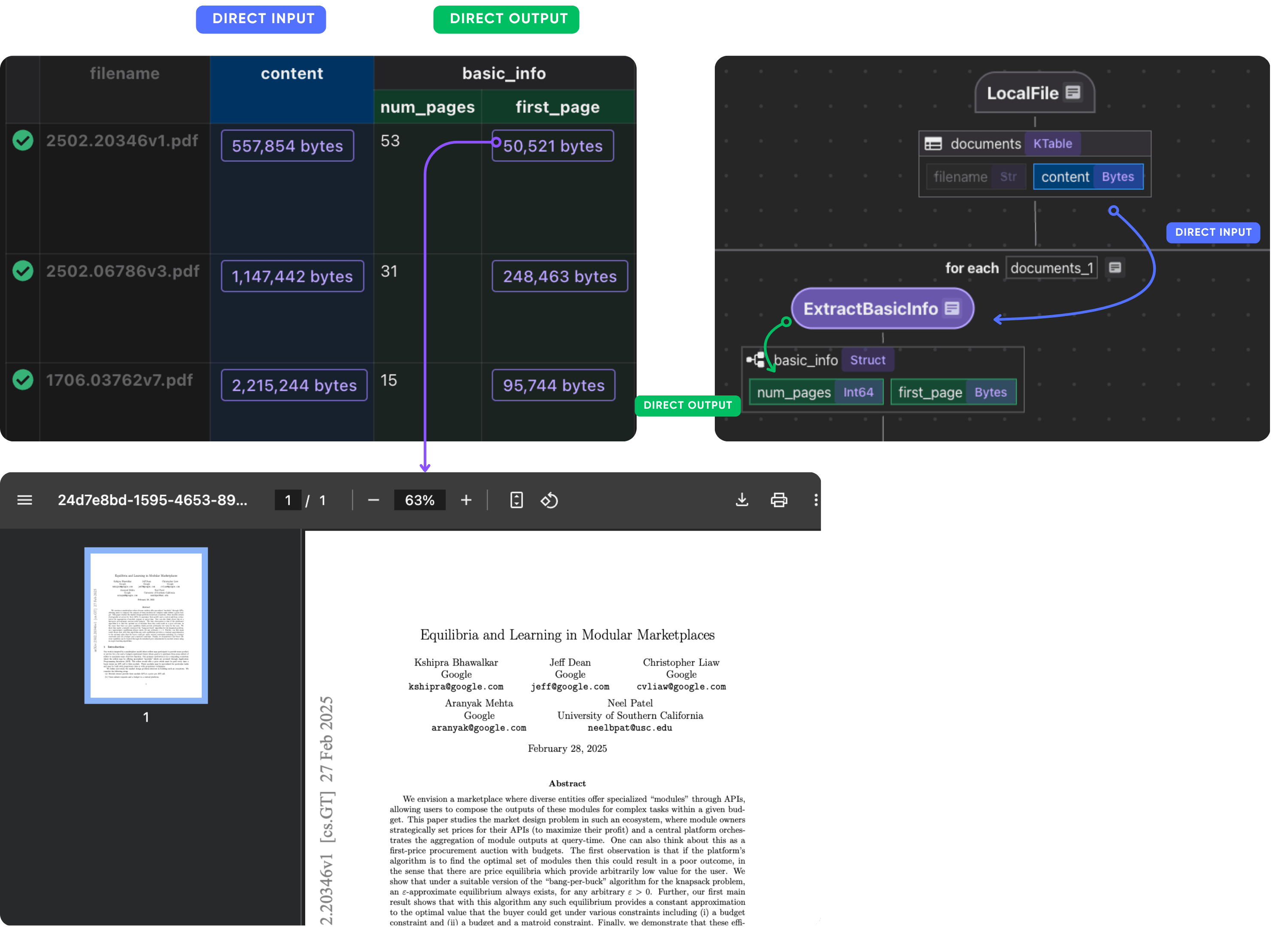
Extract first page for basic info
Define a custom function to extract the first page and number of pages of the PDF.
@dataclasses.dataclass
class PaperBasicInfo:
num_pages: int
first_page: bytes
@cocoindex.op.function()
def extract_basic_info(content: bytes) -> PaperBasicInfo:
"""Extract the first pages of a PDF."""
reader = PdfReader(io.BytesIO(content))
output = io.BytesIO()
writer = PdfWriter()
writer.add_page(reader.pages[0])
writer.write(output)
return PaperBasicInfo(num_pages=len(reader.pages), first_page=output.getvalue())
Now, plug this into your flow. We extract metadata from the first page to minimize processing cost, since the entire PDF can be very large.
with data_scope["documents"].row() as doc:
doc["basic_info"] = doc["content"].transform(extract_basic_info)
After this step, you should have the basic info of each paper.
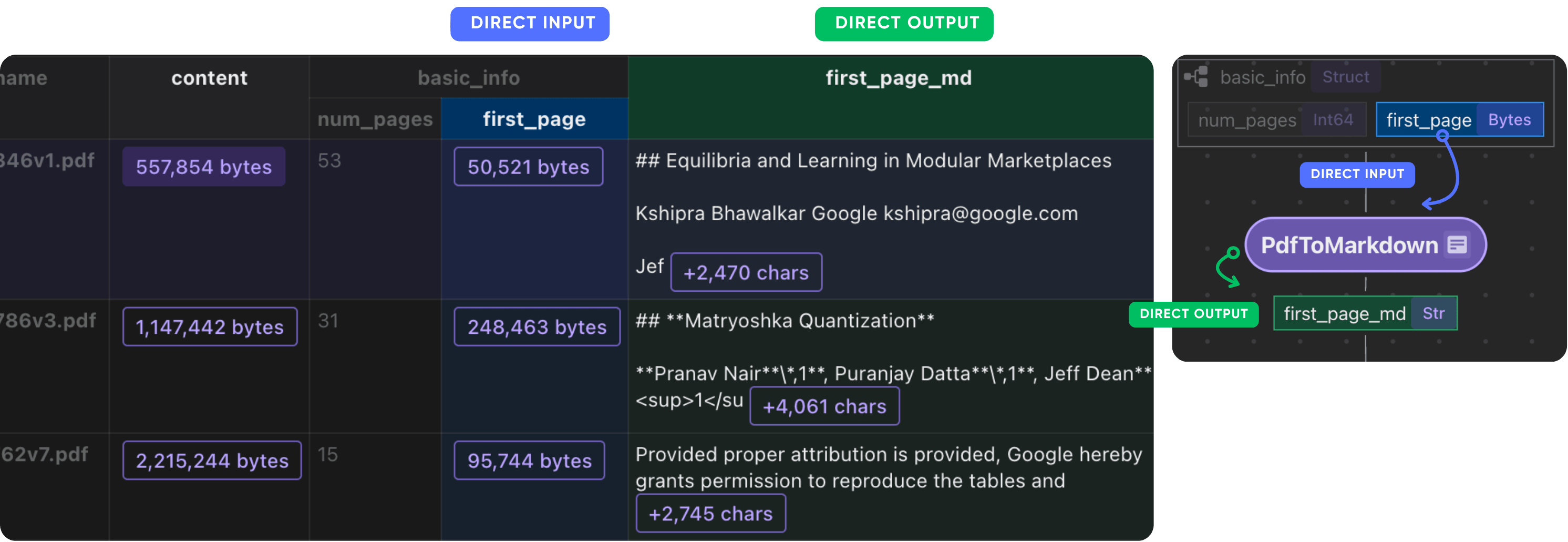
Parse basic info
We will convert the first page to Markdown using Marker. Alternatively, you can easily plug in your favorite PDF parser, such as Docling.
Define a marker converter function and cache it, since its initialization is resource-intensive. This ensures that the same converter instance is reused for different input files.
@cache
def get_marker_converter() -> PdfConverter:
config_parser = ConfigParser({})
return PdfConverter(
create_model_dict(), config=config_parser.generate_config_dict()
)
Plug it into a custom function.
@cocoindex.op.function(gpu=True, cache=True, behavior_version=1)
def pdf_to_markdown(content: bytes) -> str:
"""Convert to Markdown."""
with tempfile.NamedTemporaryFile(delete=True, suffix=".pdf") as temp_file:
temp_file.write(content)
temp_file.flush()
text, _, _ = text_from_rendered(get_marker_converter()(temp_file.name))
return text
Pass it to your transform
with data_scope["documents"].row() as doc:
doc["first_page_md"] = doc["basic_info"]["first_page"].transform(
pdf_to_markdown
)
After this step, you should have the first page of each paper in Markdown format.
Extract basic info with LLM
Define a schema for LLM extraction. CocoIndex natively supports LLM-structured extraction with complex and nested schemas. If you are interested in learning more about nested schemas, refer to this article.
@dataclasses.dataclass
class PaperMetadata:
"""
Metadata for a paper.
"""
title: str
authors: list[Author]
abstract: str
Plug it into the ExtractByLlm function. With a dataclass defined, CocoIndex will automatically parse the LLM response into the dataclass.
doc["metadata"] = doc["first_page_md"].transform(
cocoindex.functions.ExtractByLlm(
llm_spec=cocoindex.LlmSpec(
api_type=cocoindex.LlmApiType.OPENAI, model="gpt-4o"
),
output_type=PaperMetadata,
instruction="Please extract the metadata from the first page of the paper.",
)
)
After this step, you should have the metadata of each paper.
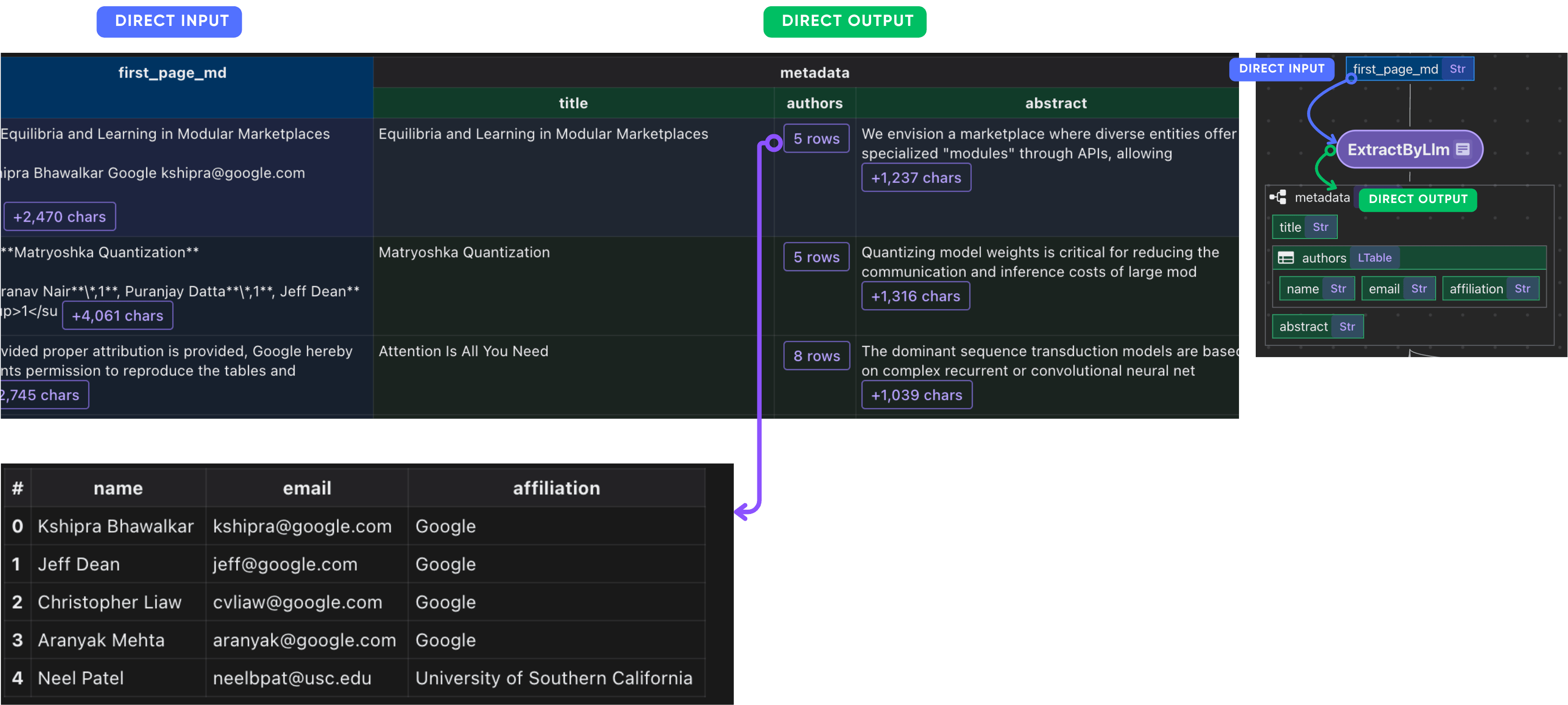
Collect paper metadata
paper_metadata = data_scope.add_collector()
with data_scope["documents"].row() as doc:
# ... process
# Collect metadata
paper_metadata.collect(
filename=doc["filename"],
title=doc["metadata"]["title"],
authors=doc["metadata"]["authors"],
abstract=doc["metadata"]["abstract"],
num_pages=doc["basic_info"]["num_pages"],
)
Just collect anything you need :)
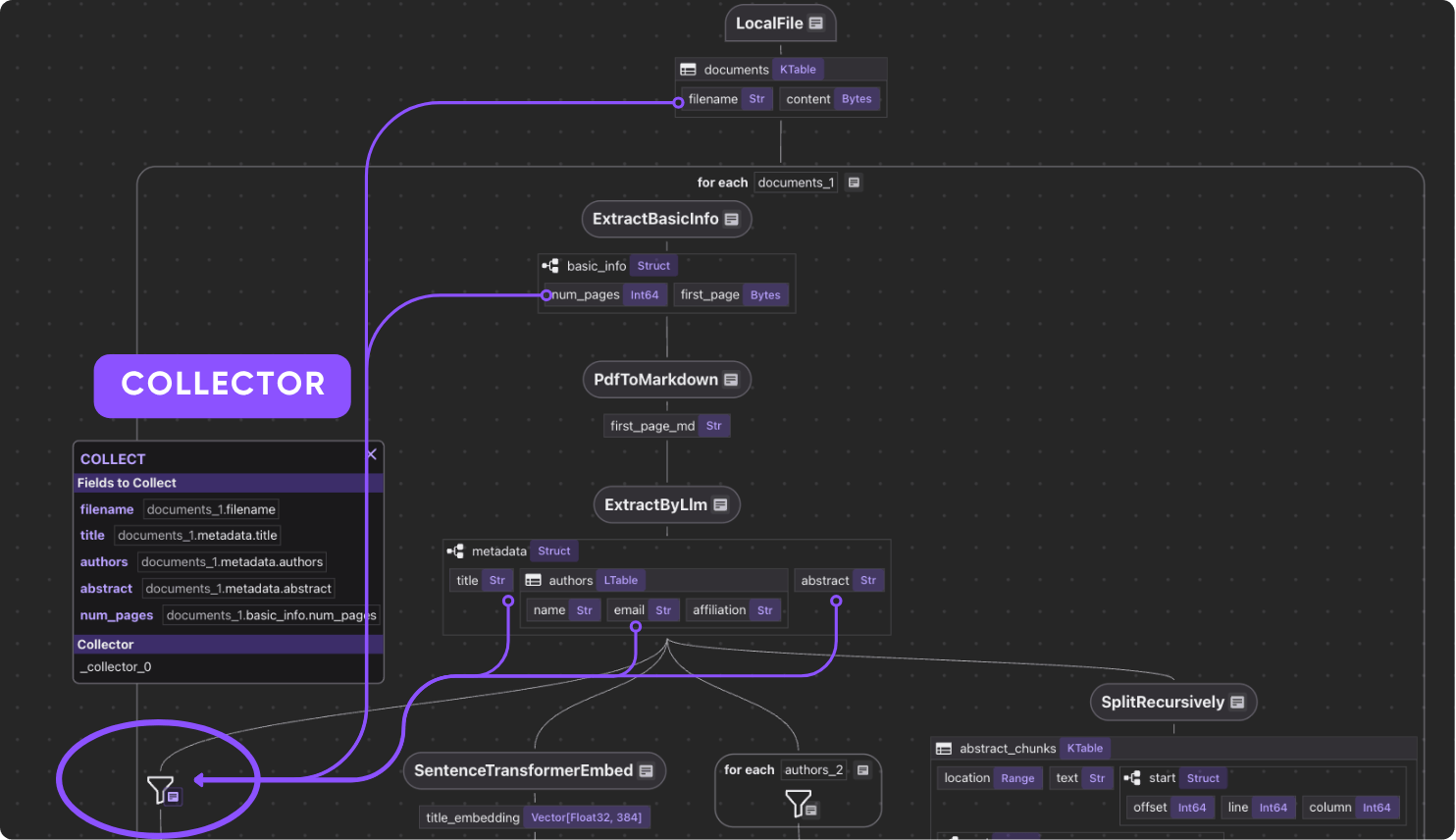
Collect author to filename information
We’ve already extracted author list. Here we want to collect Author → Papers in a separate table to build a look up functionality. Simply collect by author.
author_papers = data_scope.add_collector()
with data_scope["documents"].row() as doc:
with doc["metadata"]["authors"].row() as author:
author_papers.collect(
author_name=author["name"],
filename=doc["filename"],
)
Compute and collect embeddings
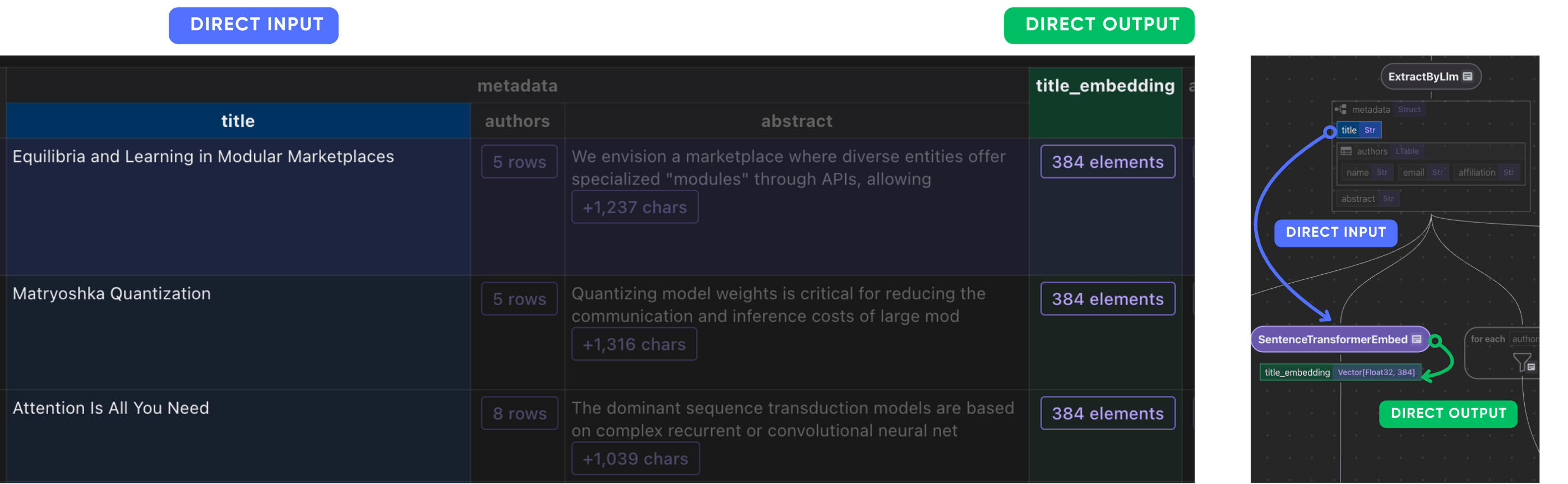
Title
doc["title_embedding"] = doc["metadata"]["title"].transform(
cocoindex.functions.SentenceTransformerEmbed(
model="sentence-transformers/all-MiniLM-L6-v2"
)
)
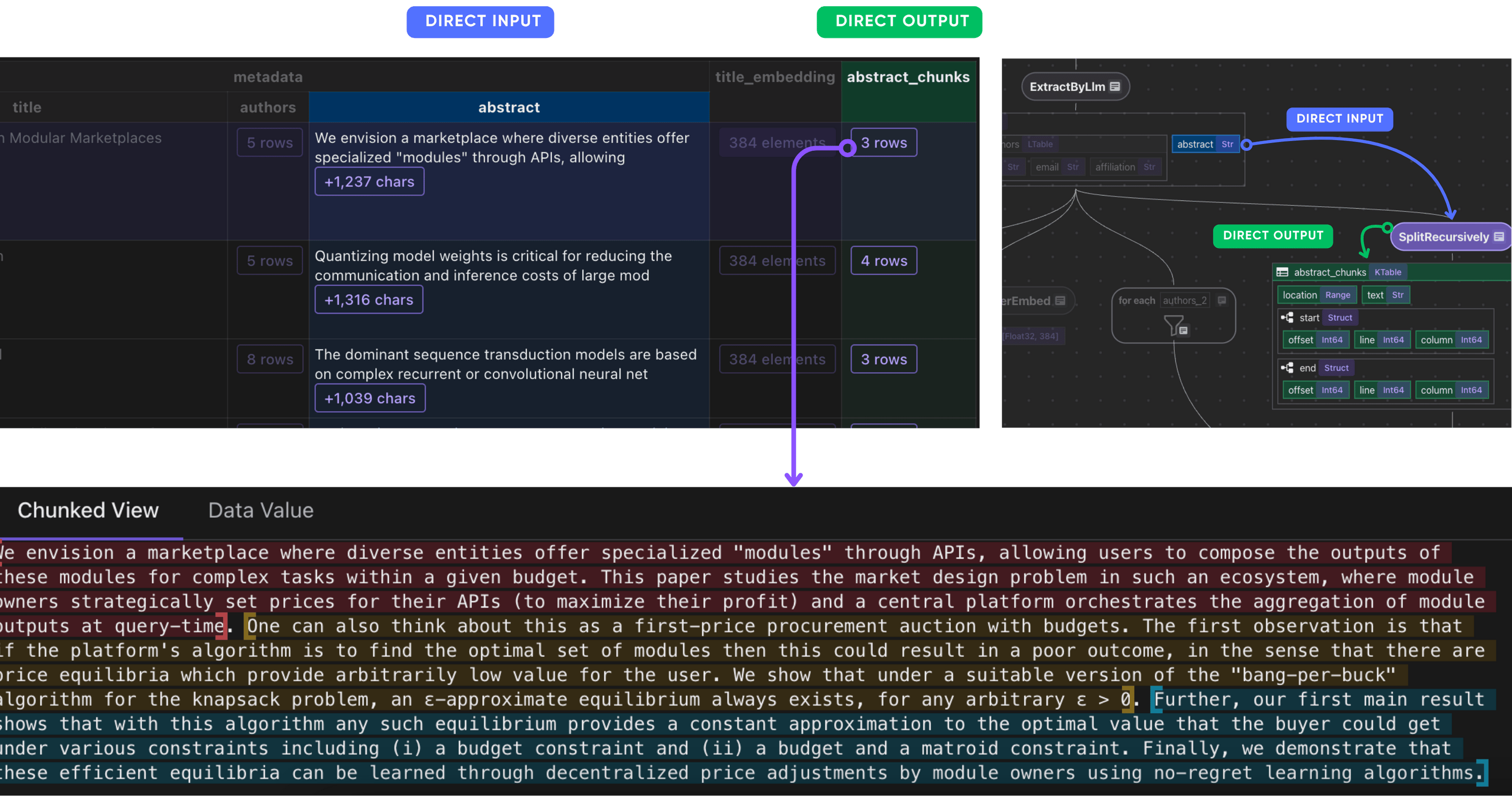
Abstract
Split abstract into chunks, embed each chunk and collect their embeddings. Sometimes the abstract could be very long.
doc["abstract_chunks"] = doc["metadata"]["abstract"].transform(
cocoindex.functions.SplitRecursively(
custom_languages=[
cocoindex.functions.CustomLanguageSpec(
language_name="abstract",
separators_regex=[r"[.?!]+\s+", r"[:;]\s+", r",\s+", r"\s+"],
)
]
),
language="abstract",
chunk_size=500,
min_chunk_size=200,
chunk_overlap=150,
)
After this step, you should have the abstract chunks of each paper.
Embed each chunk and collect their embeddings.
with doc["abstract_chunks"].row() as chunk:
chunk["embedding"] = chunk["text"].transform(
cocoindex.functions.SentenceTransformerEmbed(
model="sentence-transformers/all-MiniLM-L6-v2"
)
)
After this step, you should have the embeddings of the abstract chunks of each paper.
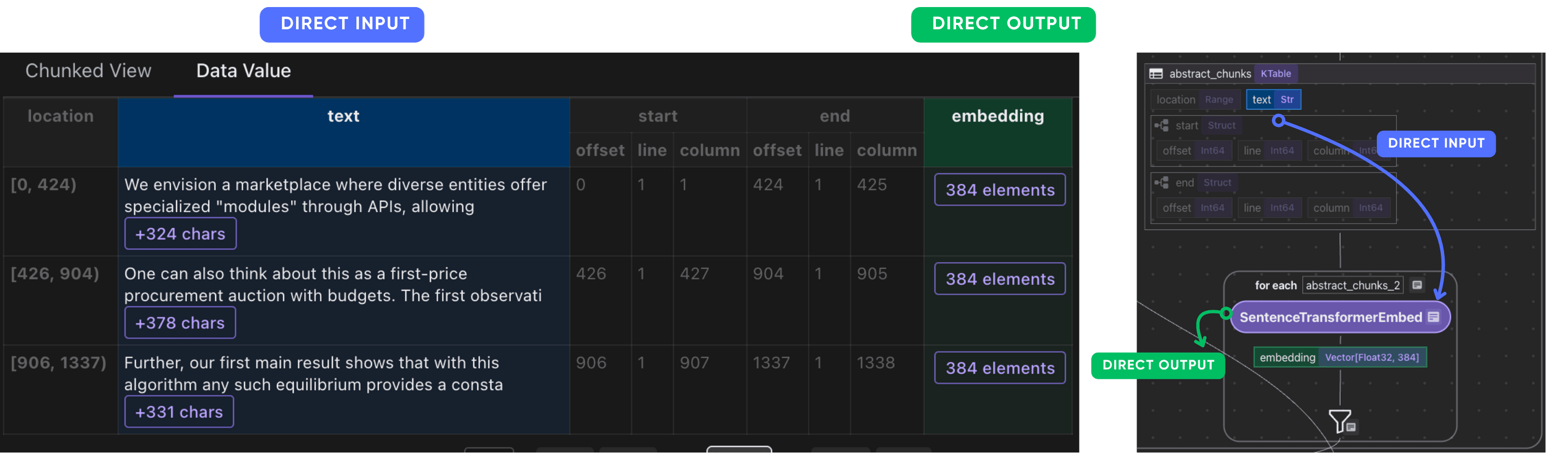
Collect embeddings
metadata_embeddings = data_scope.add_collector()
with data_scope["documents"].row() as doc:
# ... process
# collect title embedding
metadata_embeddings.collect(
id=cocoindex.GeneratedField.UUID,
filename=doc["filename"],
location="title",
text=doc["metadata"]["title"],
embedding=doc["title_embedding"],
)
with doc["abstract_chunks"].row() as chunk:
# ... process
# collect abstract chunks embeddings
metadata_embeddings.collect(
id=cocoindex.GeneratedField.UUID,
filename=doc["filename"],
location="abstract",
text=chunk["text"],
embedding=chunk["embedding"],
)
Export
Finally, we export the data to Postgres.
paper_metadata.export(
"paper_metadata",
cocoindex.targets.Postgres(),
primary_key_fields=["filename"],
)
author_papers.export(
"author_papers",
cocoindex.targets.Postgres(),
primary_key_fields=["author_name", "filename"],
)
metadata_embeddings.export(
"metadata_embeddings",
cocoindex.targets.Postgres(),
primary_key_fields=["id"],
vector_indexes=[
cocoindex.VectorIndexDef(
field_name="embedding",
metric=cocoindex.VectorSimilarityMetric.COSINE_SIMILARITY,
)
],
)
In this example we use PGVector as embedding stores/ With CocoIndex, you can do one line switch on other supported Vector databases like Qdrant, see this guide for more details. We aim to standardize interfaces and make it like assembling building blocks.
View in CocoInsight step by step
You can walk through the project step by step in CocoInsight to see exactly how each field is constructed and what happens behind the scenes.
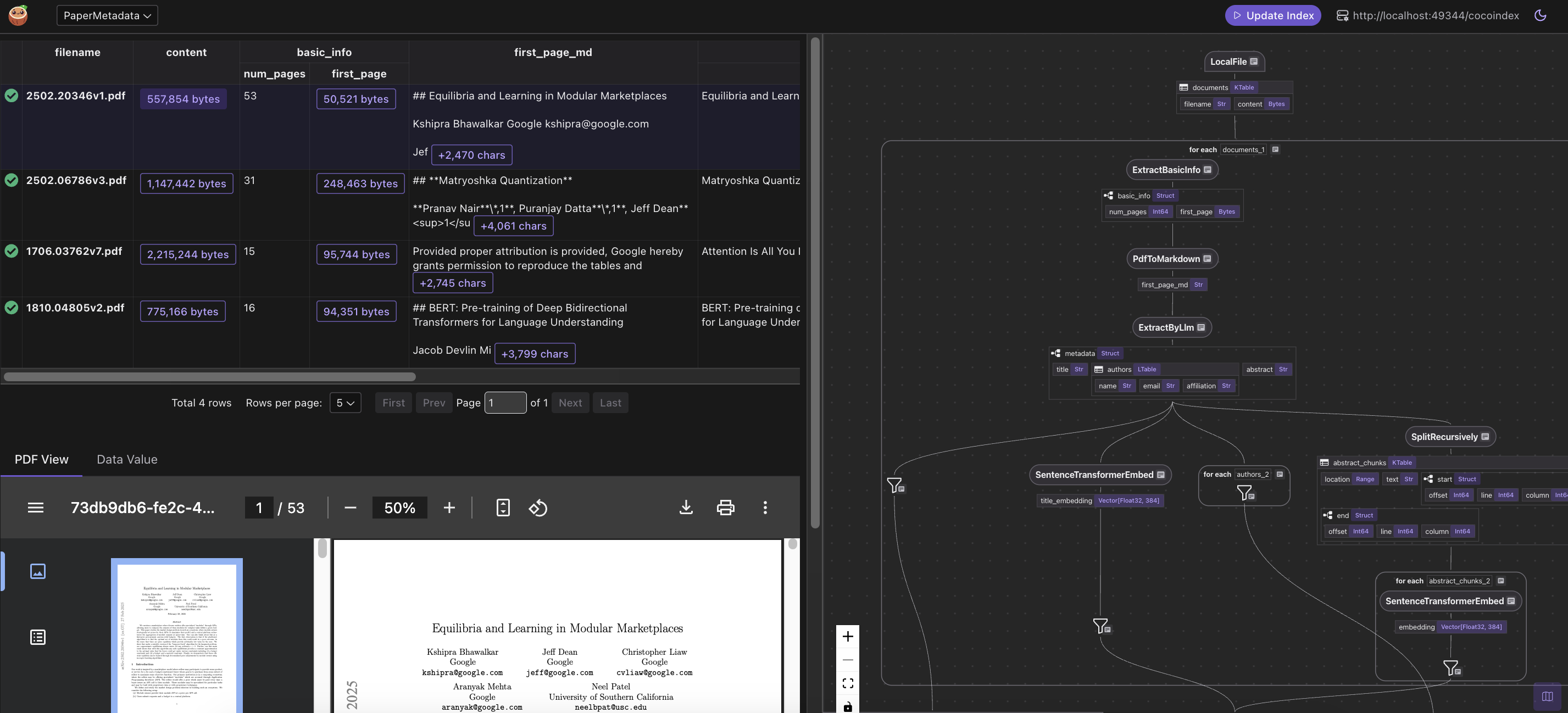
Query the index
You can refer to this section of Text Embeddings about how to build query against embeddings. For now CocoIndex doesn't provide additional query interface. We can write SQL or rely on the query engine by the target storage.
- Many databases already have optimized query implementations with their own best practices
- The query space has excellent solutions for querying, reranking, and other search-related functionality.
If you need assist with writing the query, please feel free to reach out to us at Discord.
Support us
We are constantly improving, and more features and examples are coming soon. If this article is helpful to you, please give us a star ⭐ at GitHub to help us grow.
Thanks for reading!
