Extracting Intake Forms with BAML and CocoIndex
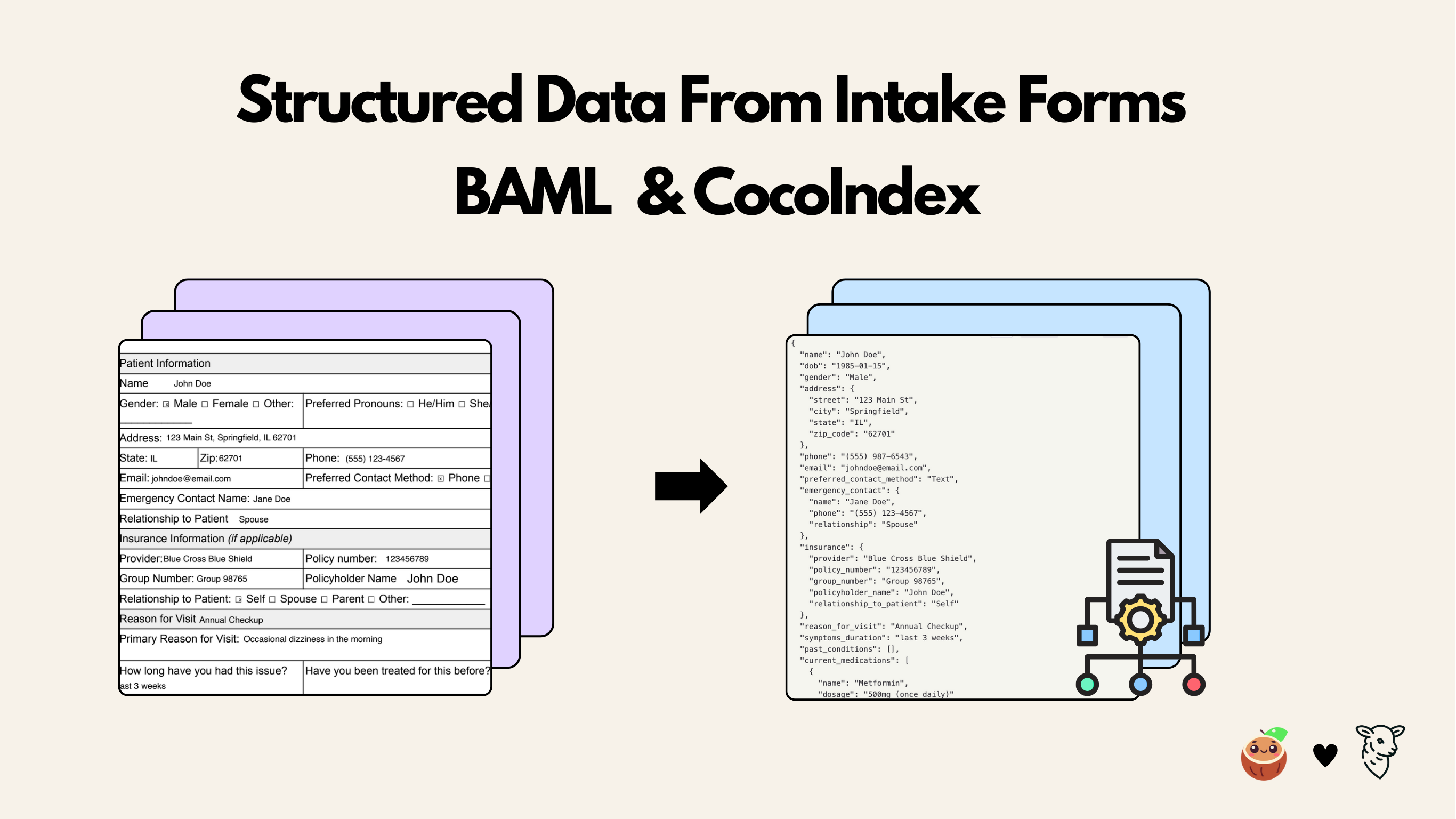
This tutorial shows how to use BAML together with CocoIndex to build a data pipeline that extracts structured patient information from PDF intake forms. The BAML definitions describe the desired output schema and prompt logic, while CocoIndex orchestrates file input, transformation, and incremental indexing.
We’ll walk through setup, defining the BAML schema, generating the Python client, writing the CocoIndex flow, and running the pipeline. Throughout, we follow best practices (e.g. caching heavy steps) and cite documentation for key concepts.
The full project is open sourced here ⭐. To see more examples build with CocoIndex, you could refer to the examples page.
BAML
BAML, created by BoundaryML, is a typed prompt engineering language that makes LLM workflows predictable, testable, and production-safe. Instead of treating prompts as fragile strings, BAML lets developers define clear input parameters, output schemas, and model configurations — transforming prompts into strongly typed functions.
CocoIndex
CocoIndex is a unified data processing engine built for AI-native applications. It lets you define transformations in one declarative workflow — then keeps everything continuously up to date with real-time, incremental processing. Designed for reliability and scale, CocoIndex ensures that every derived artifact (embeddings, metadata, extractions, models) always reflects the latest source data, making it the foundation for fast, consistent RAG, analytics, and automation pipelines.
Flow overview
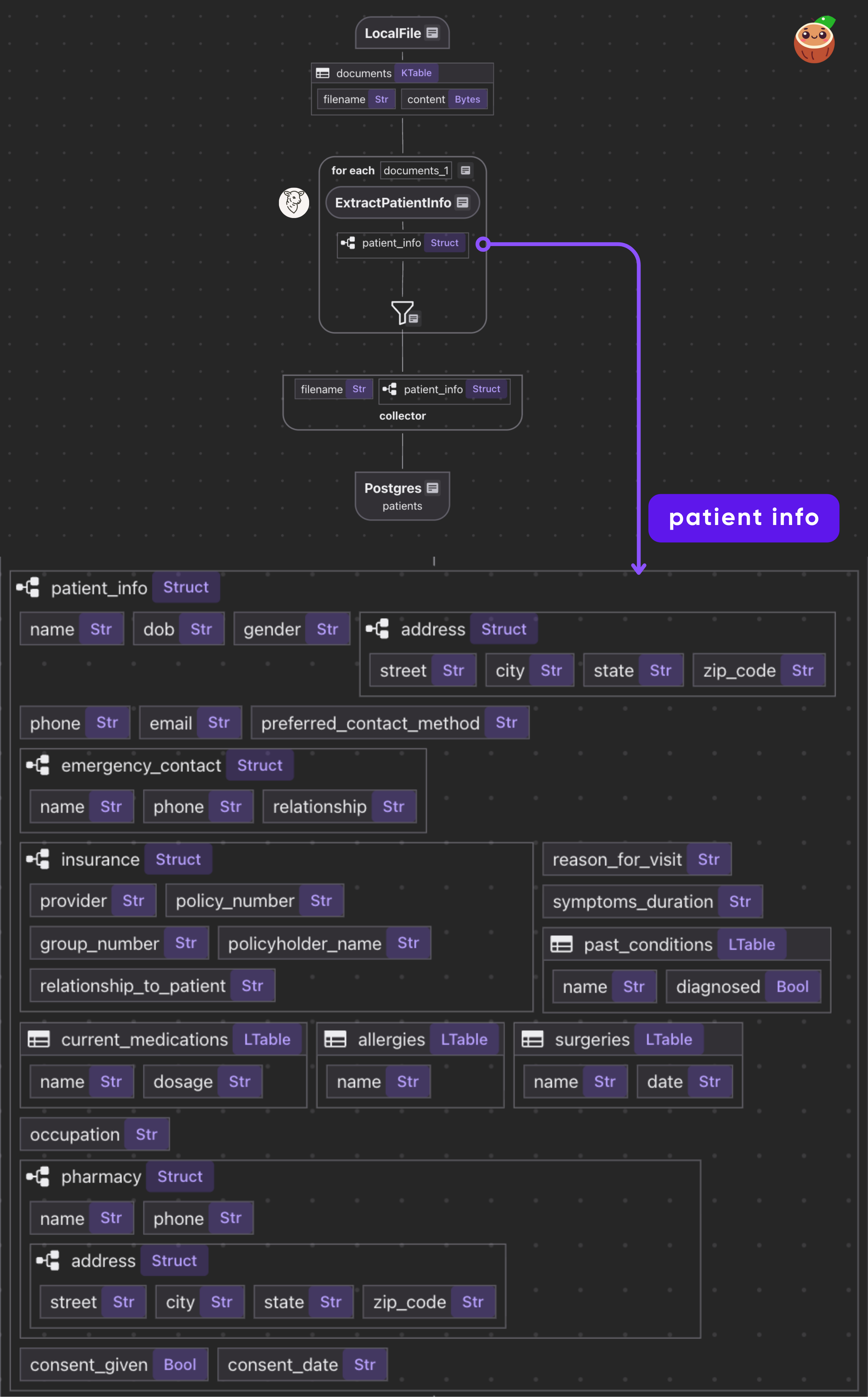
- Read PDF files from a directory.
- For each file, call the BAML function to get a structured
Patient. - Collect results and export to Postgres.
Prerequisites
-
Install Postgres if you don't have one.
-
Install dependencies
pip install -U cocoindex baml-py -
Create a
.envfile. You can copy it from.env.examplefirst:cp .env.example .envThen edit the file to fill in your
GEMINI_API_KEY.
Structured extraction component with BAML
Create a baml_src/ directory for your BAML definitions. We’ll define a schema for patient intake data (nested classes) and a function that prompts Gemini to extract those fields from a PDF. Save this as baml_src/patient.baml
Define patient schema
Classes: We defined Pydantic-style classes (Contact, Address, Insurance, etc.) to match the FHIR-inspired patient schema. These become typed output models. Required fields are non-nullable; optional fields use ?.

class Contact {
name string
phone string
relationship string
}
class Address {
street string
city string
state string
zip_code string
}
class Pharmacy {
name string
phone string
address Address
}
class Insurance {
provider string
policy_number string
group_number string?
policyholder_name string
relationship_to_patient string
}
class Condition {
name string
diagnosed bool
}
class Medication {
name string
dosage string
}
class Allergy {
name string
}
class Surgery {
name string
date string
}
class Patient {
name string
dob string
gender string
address Address
phone string
email string
preferred_contact_method string
emergency_contact Contact
insurance Insurance?
reason_for_visit string
symptoms_duration string
past_conditions Condition[]
current_medications Medication[]
allergies Allergy[]
surgeries Surgery[]
occupation string?
pharmacy Pharmacy?
consent_given bool
consent_date string?
}
Define the BAML function to extract patient info from a PDF
function ExtractPatientInfo(intake_form: pdf) -> Patient {
client Gemini
prompt #"
Extract all patient information from the following intake form document.
Please be thorough and extract all available information accurately.
{{ _.role("user") }}
{{ intake_form }}
Fill in with "N/A" for required fields if the information is not available.
{{ ctx.output_format }}
"#
}
We specify client Gemini and a prompt template. The special variable {{ intake_form }} injects the PDF, and {{ ctx.output_format }} tells BAML to expect the structured format defined by the return type. The prompt explicitly asks Gemini to extract all fields, filling “N/A” if missing.
When using BAML to extract structured data (like a Patient record) from PDFs, it is absolutely critical to ensure the PDF content is injected as part of the user message in the prompt.
Specifically, you need to include {{ _.role("user") }} before you insert your file data with {{ intake_form }}:
Why role("user") matters?
- For OpenAI models (e.g., GPT-4, GPT-4o), if the file's content is not presented in the user message, the model won't "see" the PDF at all — your extraction will fail or be empty.
- For Gemini and Anthropic, it's more forgiving and can sometimes work anyway, which makes this confusing to debug across providers.
We only discovered this after a discussion on the BAML repo and our own investigations.
If you skip the explicit role("user"), you might waste hours debugging inconsistent extractions.
Takeaway:
When building extraction flows with BAML, always set the role to "user" before adding file content to your prompt. That makes your workflow robust and portable across LLM providers.
Thanks to Deepu and Prashanth from our discord community for working with us on this issue. You can see a real-world debugging journey in our Discord thread.
Configure the LLM client to use Google’s Gemini model
client<llm> Gemini {
provider google-ai
options {
model gemini-2.5-flash
api_key env.GEMINI_API_KEY
}
}
Configure BAML generator
In baml_src folder add generator.baml:
generator python_client {
output_type python/pydantic
output_dir "../"
version "0.213.0"
}
The generator block tells baml-cli to create a Python client with Pydantic models in the parent directory.
When we run baml-cli generate
This will compile the .baml definitions into a baml_client/ Python package in your project root. It contains:
baml_client/types.pywith Pydantic classes (Patient, etc.).baml_client/sync_client.pyandasync_client.pywith a callablebobject. For example,b.ExtractPatientInfo(pdf)will return aPatient.
Continuous data transformation flow with incremental processing
Next we will define data transformation flow with CocoIndex. Once you declared the state and transformation logic, CocoIndex will take care of all the state change for you from source to target.
CocoIndex flow
Declare flow
Declare a Cocoindex flow, connect to the source, add a data collector to collect processed data.
@cocoindex.flow_def(name="PatientIntakeExtractionBaml")
def patient_intake_extraction_flow(
flow_builder: cocoindex.FlowBuilder, data_scope: cocoindex.DataScope
) -> None:
data_scope["documents"] = flow_builder.add_source(
cocoindex.sources.LocalFile(
path=os.path.join("data", "patient_forms"), binary=True
)
)
patients_index = data_scope.add_collector()
This iterates over each document. We transform doc["content"] (the bytes) by our extract_patient_info function. The result is stored in a new field patient_info. Then we collect a row with the filename and extracted patient info.
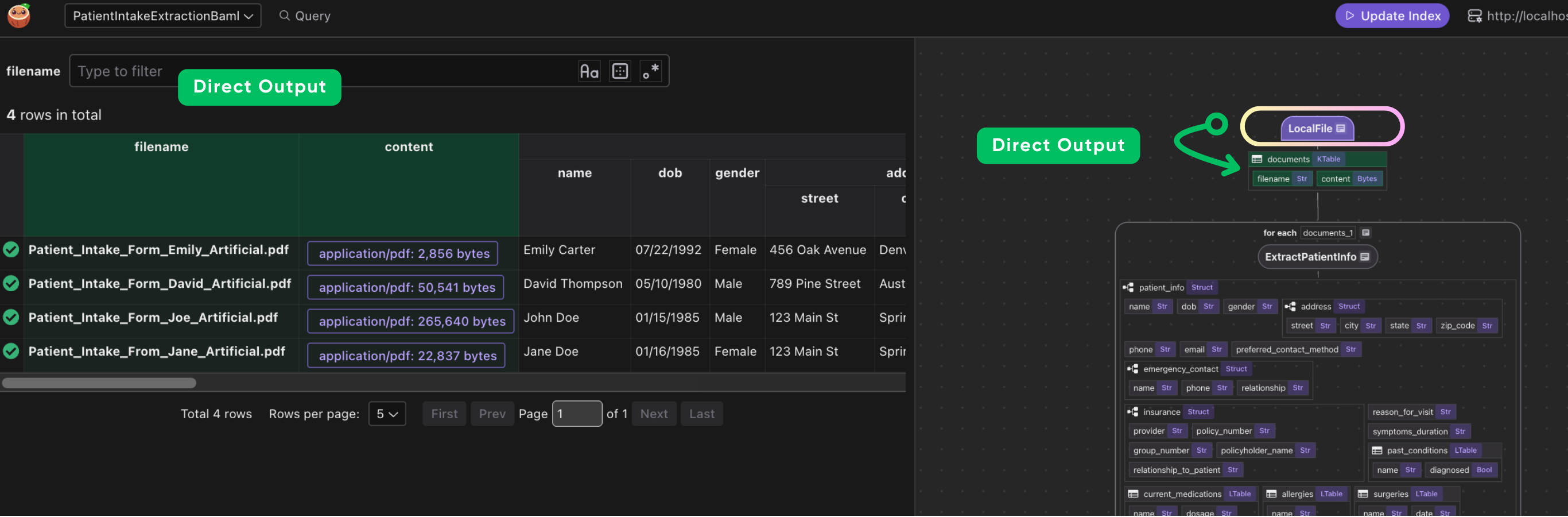
Define a custom function to use BAML extraction to transform a PDF
@cocoindex.op.function(cache=True, behavior_version=1)
async def extract_patient_info(content: bytes) -> Patient:
pdf = baml_py.Pdf.from_base64(base64.b64encode(content).decode("utf-8"))
return await b.ExtractPatientInfo(pdf)
- The
extract_patient_infofunction is decorated with@cocoindex.op.function(cache=True, behavior_version=1). Settingcache=Truecauses CocoIndex to cache outputs of this function for incremental runs (so unchanged inputs skip rerunning the LLM). We increasebehavior_version(start at 1) so that any prompt or logic changes will force a refresh. - Inside the function, we convert
bytesto a BAMLPdf(via base64) and then callawait b.ExtractPatientInfo(pdf). This returns aPatientdataclass instance (mapped from the BAML output)
Process each document
- Transform each doc with BAML
- collect the structured output
with data_scope["documents"].row() as doc:
doc["patient_info"] = doc["content"].transform(extract_patient_info)
patients_index.collect(
filename=doc["filename"],
patient_info=doc["patient_info"],
)
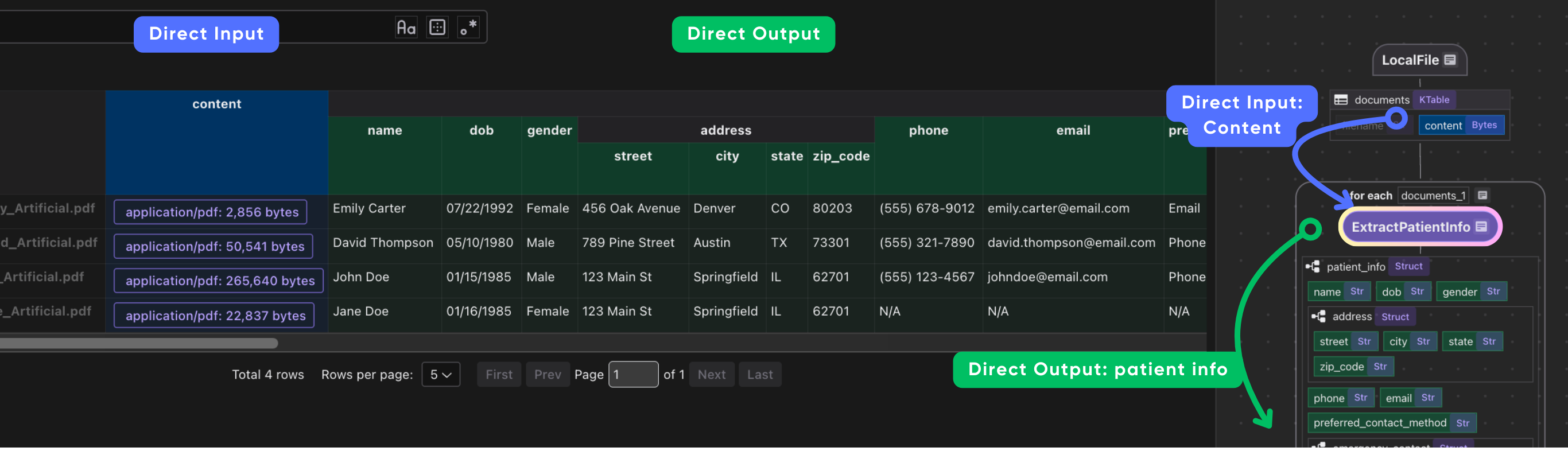
It is common to have heavy nested data, CocoIndex is natively designed to handle heavily nested data structures.
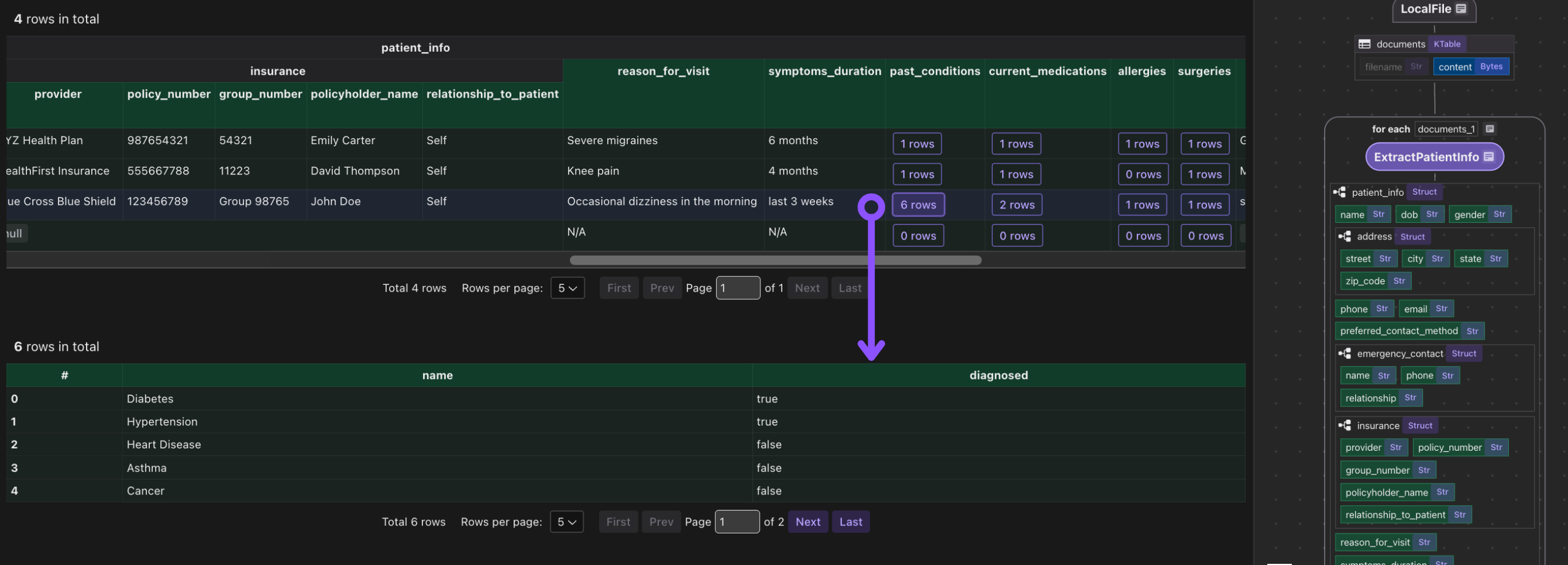
Export to Postgres
patients_index.export(
"patients",
cocoindex.storages.Postgres(),
primary_key_fields=["filename"],
)
we export the collected index to Postgres. This will create/maintain a table patients keyed by filename, automatically deleting or updating rows if inputs change. Because CocoIndex tracks data lineage, it will handle updates/deletions of source files incrementally
Running the pipeline
Generate BAML client code (required step, in case you didn’t do it earlier. )
baml generate
This generates the baml_client/ directory with Python code to call your BAML functions.
Update the index:
cocoindex update main
CocoInsight
I used CocoInsight (Free beta now) to troubleshoot the index generation and understand the data lineage of the pipeline. It just connects to your local CocoIndex server, with zero pipeline data retention.
cocoindex server -ci main
Composable by default: use the best components for your use case
While CocoIndex provides a rich set of building blocks for building LLM pipelines, it is fundamentally designed as an open system. Developers can bring in their preferred transformation components tailored to their domain — from document parsers to structured extractors like BAML.
This flexibility enables deep composability with other open ecosystems. The synergy between CocoIndex and BAML highlights this philosophy: BAML brings powerful prompt-driven schema extraction, while CocoIndex orchestrates and maintains the flow at scale. There’s no lock-in — developers and enterprises experimenting at the frontier can adapt, extend, and integrate freely.
Summary
By combining BAML and CocoIndex, we get a robust, schema-driven workflow: BAML ensures the prompt-to-schema mapping is correct and type-safe, while CocoIndex handles data ingestion, transformation, and incremental storage. This example extracted patient intake information (names, insurance, medications, etc.) from PDFs, but the pattern applies to any structured data extraction task.
- For more details on CocoIndex flows, see its documentation.
- For BAML usage and prompt engineering, refer to the BoundaryML docs.
